Large language models (LLMs) often struggle with accuracy when handling domain-specific questions, especially those requiring multi-hop reasoning or access to proprietary data. While retrieval-augmented generation (RAG) can help, traditional vector search methods often fall short.
In this tutorial, we show you how to implement GraphRAG in combination with fine-tuned GNN+LLM models to achieve 2x accuracy compared to standard baselines.
This approach is particularly valuable for scenarios involving:
- Domain-specific knowledge (product catalogs, supply chains)
- High-value proprietary information (drug discovery, financial models)
- Privacy-sensitive data (fraud detection, patient records)
How GraphRAG works
This specific approach of graph-powered retrieval-augmented generation (GraphRAG) builds on the G-Retriever architecture. G-Retriever represents grounding data as a knowledge graph combining graph-based retrieval with neural processing:
- Knowledge graph construction: Represent domain knowledge as a graph structure.
- Intelligent retrieval: Use graph queries and the Prize-Collecting Steiner Tree (PCST) algorithm to find relevant subgraphs.
- Neural processing: Integrate GNN layers during LLM fine-tuning to optimize attention on retrieved context.
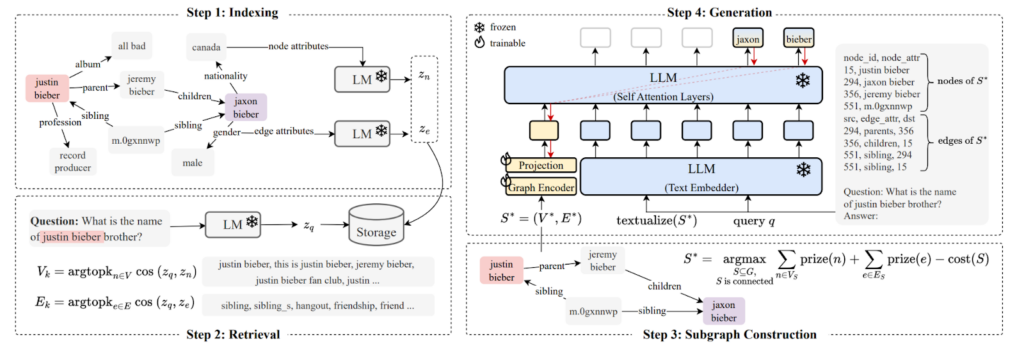
(source: G-Retriever: Retrieval-Augmented Generation for Textual Graph Understanding and Question Answering)
The process works with the training data triplets {(Qi, Ai, Gi)}:
- Qi: Multi-hop natural language question
- Ai: Set of answer nodes
- Gi = (Vi, Ei): Relevant subgraph (already obtained by some method)
The pipeline follows these steps:
- Find semantically similar nodes
and edges
to question Qi.
- Assign high prizes to these matching nodes and edges.
- Run a variant of the PCST algorithm to find the optimal subgraph
that maximizes prize while minimizing size.
- Fine-tune combined GNN+LLM model on {(Qi, Gi*)} pairs to predict {Ai}.
PyG offers a modular setup for G-Retriever. Our code repository integrates this with a graph database for persisting large graphs and a vector index as well as providing the retrieval query templates.
Real-world example: Biomedical Q&A
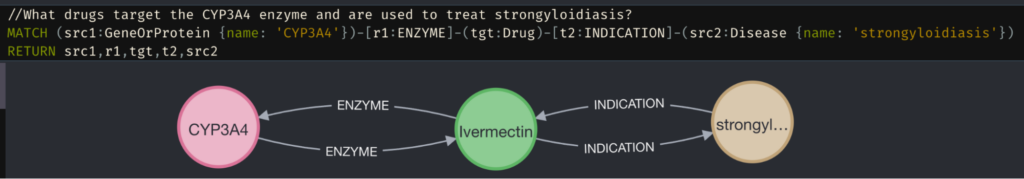
Look at the STaRK-Prime biomedical dataset. Consider the question, “What drugs target the CYP3A4 enzyme and are used to treat strongyloidiasis?”
The correct answer (Ivermectin) requires understanding the following:
- Direct relationships (drug-enzyme, drug-disease connections)
- Node properties (drug descriptions and classifications)
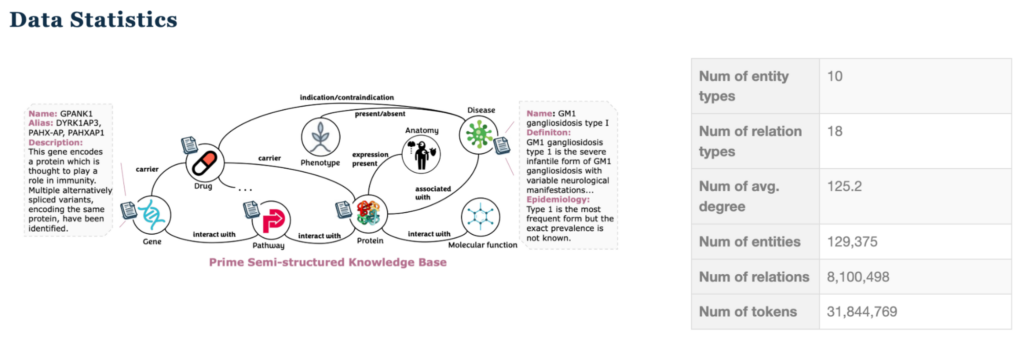
This dataset is particularly challenging due to the following factors:
- Heterogeneous node and relationship types
- Variable-length text attributes
- High average node degree causing neighborhood explosion
- Complex multi-hop reasoning requirements
Implementation details
To follow along with this tutorial, we recommend familiarity with the following:
- Graph databases: Working knowledge of Neo4j and Cypher queries
- Graph neural networks (GNNs): Basic usage of PyTorch Geometric (PyG)
- LLMs: Experience with model fine-tuning
- Vector search: Understanding of embeddings and similarity search
For more information about graph databases and GNNs, see the Neo4j documentation and PyG examples. All code is available in the /neo4j-product-examples GitHub repo.
Data preparation
The benchmark dataset consists of serialized .pt files. Preprocess the files and load them into a Neo4j database as shown in stark_prime_neo4j_loading.ipynb in the GitHub repo.
Add database constraints to ensure the quality of the data, with CREATE CONSTRAINT to ensure that nodeId is unique for every node label.
CREATE CONSTRAINT unique_geneorprotein_nodeid IF NOT EXISTS FOR (n:GeneOrProtein) REQUIRE n.nodeId IS UNIQUE |
You also load a text embedding property on every node, generated by embedding the textual description with OpenAI text-embedding-ada-002.
Then, create a vector index on the text embedding with cosine similarity to speed up query time when looking for semantically similar nodes by using CREATE VECTOR INDEX.
For more information about indexing and constraints in Neo4j, see the Neo4j Cypher manual.
Subgraph retrieval
The initial retrieval process is carried out in the following steps:
- Embed incoming questions using text-embedding-ada-002 using the Langchain OpenAI API.
- Find the top four similar nodes from the database (
db.index.vector.queryNodes) using vector search. - Expand one hop from those four nodes, which gives you a large subgraph (due to the density) to create base subgraph Gi.
- Identify the top 100 relevant nodes in the base subgraph Gi using vector similarity and assign prizes (4.0 to 0.0) with a constant interval of 0.04.
- Project the graph into Neo4j GDS and run the variant of the PCST algorithm to produce a pruned subgraph Gi*.
- Run the PCST algorithm to get pruned subgraph Gi*.
For each pruned subgraph, follow these steps:
- Enrich it with node name and description as
node_attr. - Edit the question to be a prompt template of
f"Question: {question}\nAnswer: ". - Convert all answer nodeId values in the training and validation dataset to their corresponding node names.
- Concatenate them with the separator symbol
|.
Prepare a textual description of the pruned subgraph following the recipe of G-Retriever. In addition, order the nodes in the textual description as an implicit signal to the fine-tuning model. Here’s an example prompt:
node_id,node_attr14609,"name: Zopiclone, description: 'Zopiclone is...'"14982,"name: Zaleplon, description: 'Zaleplon is...'"...src,edge_attr,dst15570,SYNERGISTIC_INTERACTION,1544114609,SYNERGISTIC_INTERACTION,15441...Question: Could you suggest any medications effective for treating beriberi that are safe to use with Zopiclone?\nAnswer: |
At the end of this process, you have a set of training PyTorch data objects that contains the question, answers as concatenated node names, and edge_index (representing the topology of the pruned subgraph), as well as textualized description. The nodes can already be used as answers, so you can evaluate metrics such as hits@1 and recall, which are shown in the results later in this post.
To improve the metric further, you can also fine-tune a GNN+LLM.
GNN+LLM fine-tuning
Fine-tune a GNN and LLM jointly using PyG’s LLM and GNN modules, similar to the G-Retriever example:
- GNN: GATv1 (Graph Attention Network)
- LLM: meta-llama/Llama-3.1-8B-Instruct
- Training: 6K Q&As on four A100 40GB GPUs (~2 hours)
- Context length: 128k tokens to handle long node descriptions
The choice of Llama model is due to its 128k context length, which enables you to handle long textual node descriptions without restricting the subgraph size.
Fine-tuning the GNN+LLM model on four A100 40GB GPUs takes about 2 hours to train 6K Q&As and evaluate 4K Q&As. This process scales linearly with the number of training examples.
Results
This approach achieves significant improvements (Table 1):
| Method | Hits@1 | Hits@5 | Recall@20 | MRR |
| Pipeline* | 32.09 | 48.34 | 47.85 | 38.48 |
| G-Retriever | 32.27±0.3 | 37.92±0.2 | 27.16±0.1 | 34.73±0.3 |
| Subgraph Pruning** | 12.60 | 31.60 | 35.93 | 20.96 |
| Baseline | 15.57 | 33.42 | 39.09 | 24.11 |
**Frozen LLM. No fine-tuning.
Table 1. Results of our methods compared to the baseline across 4 different metrics.
Here are some of our key findings:
- 32% hits@1: More than double the baseline.
- Pipeline approach: Combines the strengths of both pruned subgraphs and G-Retriever.
- Sub-second inference: For real-world queries.
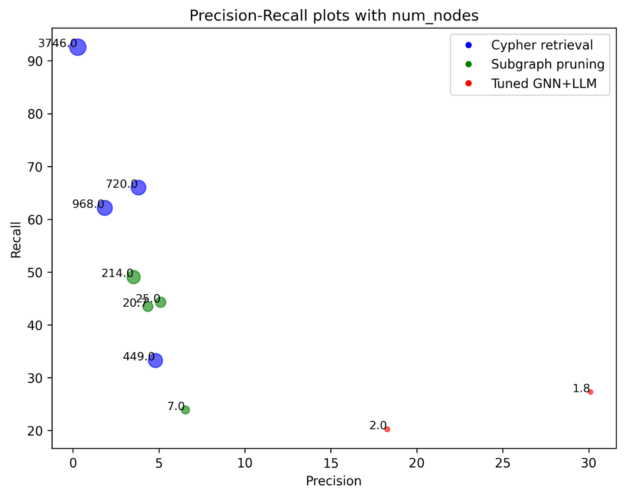
Our result obtains 32% hits@1, which is more than double the reported baseline from STaRK-Prime. The pruned subgraph produced in the intermediate step also already achieves scores close to the best baseline without any fine-tuning of GNN+LLM.
However, the pruned subgraph contains many more nodes, whereas the output from G-Retriever usually only has 1–2 nodes. This is expected as, on average, the ground-truth answers have 1–2 nodes.
Hits@5 and Recall@20 is therefore an unfair metric to G-Retriever, when it does not produce even 5 answers. It is fine-tuned to return exactly the right answers. On the other hand, hits@1 is an unfair metric for pruned subgraphs, as there is no constraint on the ordering of the nodes in the pruned subgraph.
Therefore, append to the G-Retriever output any unique nodes from the pruned subgraph, which was the input to G-Retriever, up to 20 nodes. We denote this simple ensemble model as the pipeline. Scores obtained by this pipeline are significantly higher than the best baseline by a wide margin across all metrics.
During runtime, given a question, obtaining the base subgraph, running PCST for a pruned subgraph, the forward pass of the GNN+LLM and all the intermediate steps can finish within seconds.
| Time(s)/Query | Min | Median | Max |
| Cypher | 0.056 | 0.069 | 1.179 |
| PCST | 0.044 | 0.166 | 3.573 |
| GNN+LLM | 0.410 | 0.497 | 0.562 |
Challenges and future work
There are still several challenges and limitations to the existing methods, such as:
- Hyperparameter complexity
- Large discrete search space (for example, the number of hops in the Cypher expansion, node and relationship filtering, node and edge prize assignments, and so on)
- Multiple interconnected parameters affecting performance
- Difficult to find optimal configurations
- Dataset challenges
- Handling polysemous/synonymous terms
- All current benchmarks are limited to ≤4 hop questions
- Assumption that answers are nodes rather than subgraphs
- Assumption of complete (no missing edges) graphs
The following example shows a question where the correct answer node 62592 is hyaluronan metabolism, while our model finds 47408 (hyaluronan metabolic process). It’s hard to tell what the difference is and why one node is preferred over the other as the true answer.
Q: Please find a metabolic process that either precedes or follows hyaluronan absorption and breakdown, and is involved in its metabolism or signaling cascades.Label (Synthesized): hyaluronan metabolism (id: 62592)Our answer ("Incorrect"): hyaluronan metabolic process (id: 47408)+ hyaluronan catabolic process, absorption of hyaluronan, hyal |
Future directions
We point to the readers several promising future directions that we believe will improve GraphRAG further:
- Advanced architecture
- Explore graph transformers
- Support global attention
- Scale to handle larger hop distances
- Support complex subgraph answers
- Robustness
- Handle incomplete or noisy graphs
- Improve disambiguation of similar concepts
- Scale to enterprise-scale knowledge graphs
For more information, see the following resources:
- /neo4j-product-examples/neo4j-gnn-llm-example GitHub repo, with all the code to replicate and directions for setup
- graphrag.com and What is GraphRAG for more information about GraphRAG and graph construction patterns
- GNN+LLM Stanford Lecture