NVIDIA Parabricks is a scalable genomics analysis software suite that solves omics challenges with accelerated computing and deep learning to unlock new scientific breakthroughs.
Released at NVIDIA GTC 2025, NVIDIA Parabricks v4.5 supports the growing quantity of data by including support for the latest NVIDIA GPU architectures, and improved alignment and variant calling with the combination of Giraffe and DeepVariant. The release also includes improved functionality and reduced analysis time across multiple industry-leading tools, including STAR, FQ2BAM, and Minimap2.
Parabricks v4.5 is accompanied by new NVIDIA AI Blueprints for genomics and single-cell analysis, enabling bioinformaticians and genomics platform providers to easily deploy and test NVIDIA Parabricks and NVIDIA RAPIDS without requiring local GPUs or self-managed cloud provisions. By expanding accessibility, scientists and researchers can get up and running faster to further genomic understanding.
Release highlights include the following:
What’s new
- NVIDIA Blueprints available through NVIDIA Brev, including genomics analysis using Parabricks and single-cell analysis leveraging RAPIDS and RAPIDS-singlecell.
- Parabricks support for NVIDIA Blackwell, including the new NVIDIA RTX PRO 6000 Blackwell Server Edition GPU, bringing overall germline analysis down to 7 minutes, 56 seconds using four GPUs.
- The ability to easily combine Giraffe and DeepVariant, combining the power of pangenome analysis with the gold standard for variant calling.
- Collaboration with Roche Sequencing.
Improved features
- STAR acceleration (2x acceleration over existing speed)
- Faster FQ2BAM (recovery mode improvements)
- Faster force-calling mode in Haplotypecaller/Mutectcaller
- Faster Giraffe (over 3.7x acceleration than baseline VG Giraffe)
- Minimap2 acceleration (including splice-alignment)
Easily deploy and run genomics and single-cell analysis with NVIDIA Blueprints
AI Blueprints provide reference workflows for agentic and generative AI, so enterprises can build, test, and deploy custom AI applications. Before deploying a large-scale enterprise blueprint, bioinformaticians now have access to lightweight blueprints through NVIDIA Brev, which make it easy to evaluate accelerated genomics libraries enabled by NVIDIA technologies without infrastructure setup or arduous deployments.
At NVIDIA GTC 2025, NVIDIA launched two blueprints for genomics use cases: genomics analysis and single-cell analysis. Both enable bioinformaticians and genomics platform providers to easily deploy and gain access to NVIDIA technology—including NVIDIA Parabricks and RAPIDS.
For genomics analysis, bioinformaticians can try a whole-exome sequencing analysis workflow on short reads in a matter of minutes on any cloud available through NVIDIA Brev, using NVIDIA Parabricks FQ2BAM for alignment and DeepVariant for variant calling.
For single-cell analysis, scientists can also test near-real-time data analysis by trying rapids-singlecell, developed by scverse. This will enable 676x faster UMAP and 70x faster PCA on a 1M cell dataset.
“Enabling seamless access to RAPIDS-singlecell notebooks across cloud platforms provides researchers with a powerful, user-friendly tool,” says dual-PhD Prof. Dr. Fabian J. Theis, from the School of Computation, Information and Technology and TUM School of Life Sciences, TU Munich. “By making advanced computational resources more accessible, we are fostering greater adoption of scverse tools and empowering the community to accelerate analyses and innovation in single-cell analysis.”
Improve performance with Parabricks on NVIDIA Blackwell
Advancing generative AI and accelerated computing, NVIDIA Blackwell is the latest GPU architecture that dramatically improves performance and efficiency at scale.
With the latest v4.5 release, Parabricks is now available for use with Blackwell, enabling further acceleration and reduced runtimes, leveraging increased ALU units and tensor cores for better performance. On a single RTX PRO 6000, fastq2bam and Deepvariant workflows were 1.75x faster than NVIDIA H100 PCIe.

“NVIDIA Parabricks already delivers significant speedups on our current cloud-based GPU architecture,” says Abhimanyu (Abhi) Verma, chief technology officer at SOPHiA GENETICS. “With its latest support on Blackwell, we are excited to bring even greater acceleration and enhanced performance within the SOPHiA DDM Platform. This advancement enables us to continue to provide rapid turnaround time for AI-driven analyses, empowering labs, researchers, and biopharma worldwide. The timing is ideal, as we see an exponential increase in data volumes driven by deeper sequencing and more advanced multimodal approaches across the board.”
Improve runtimes and enhance accuracy by combining Giraffe and DeepVariant
Alignment and variant calling are critical steps in analyzing genomic data that work together to help understand genetic variations. The first step occurs by arranging and comparing sequences in alignment. Next, scientists identify variants of whole genomes, exomes, and gene panels in variant calling.
Although alignment and variant calling are crucial components of high-throughput sequencing, they’re time-intensive processes that require significant computational resources—particularly when it comes to whole-genome sequencing.
To help address these bottlenecks, specialized algorithms and tools have been developed across alignment and variant calling steps—including Giraffe for alignment and DeepVariant for variant calling. Giraffe is a software tool developed by the University of California, Santa Cruz for pangenome graph alignment. DeepVariant, a deep-learning–based variant caller developed by Google. Not only are these both popular open-source tools, but they help expedite their respective processes while ensuring higher accuracy.
Both Giraffe and DeepVariant are already supported in Parabricks, but the latest v4.5 provides further acceleration by improving performance across tools individually and together.
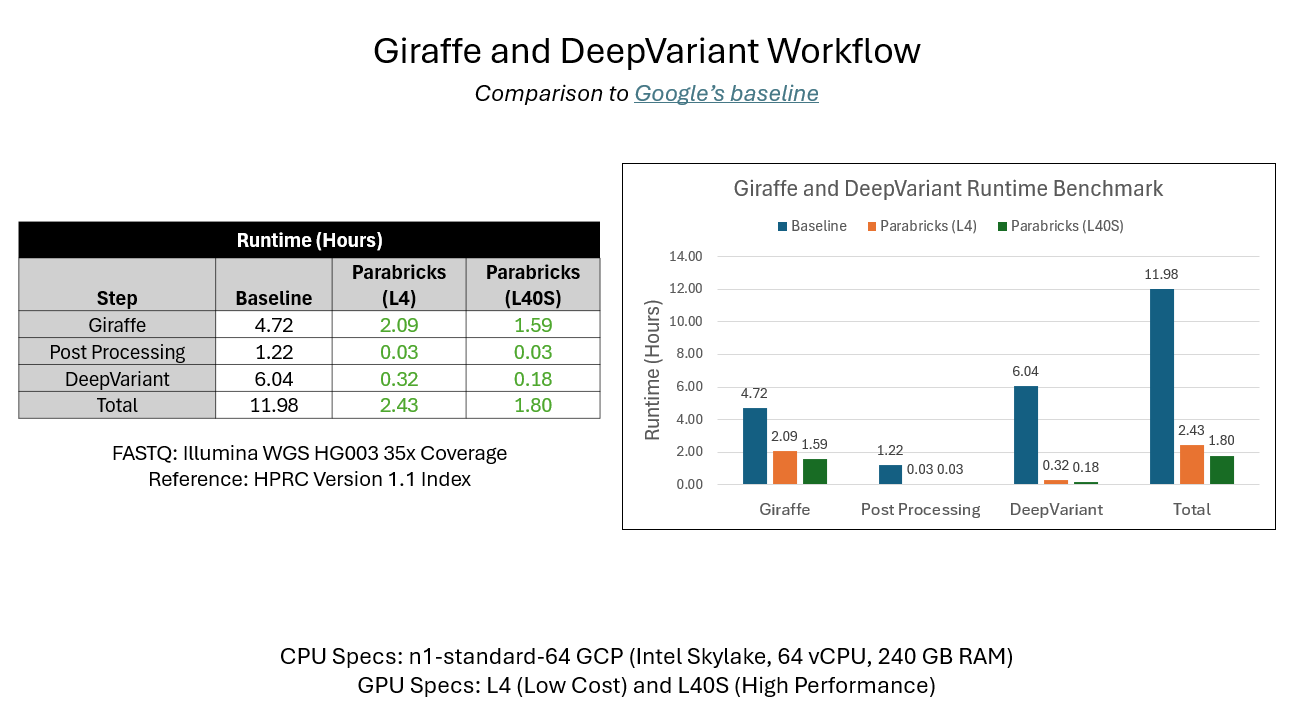
As a result, scientists and researchers can perform critical steps even faster while maintaining accuracy. For example, on NVIDIA L40S runtime is more than 6x faster compared to CPU. The total runtime for Giraffe, post-processing, and DeepVariant on CPU was 11.98 hours. On NVIDIA L40S, Parabricks v4.5 saw total runtime decrease to only 1.8 hours.
New collaboration with Roche Sequencing
Genomic data generation is at an inflection point where new technologies, like sequencing by expansion (SBX), recently unveiled by Roche, enable sequencing at unparalleled rates, requiring faster, higher throughput analysis.
Along with their proprietary accelerated algorithms, Roche has integrated NVIDIA hardware and software components directly into the SBX platform—including select Parabricks components and other accelerated libraries. Roche’s work on accelerated algorithms, in collaboration with NVIDIA, will provide an advanced data analysis architecture that plays a key role in realizing the full potential of the ultra-rapid data generation rates and fast sequencing run times enabled by SBX.
“SBX technology offers significant advantages through high sequencing data generation rates and flexible run times, enhanced by real-time, integrated analysis,” says John Mannion, VP Computational Sciences at Roche. “The strategic collaboration with NVIDIA is an important component of Roche’s accelerated pipeline development, enabling efficient local analysis of compute-heavy data processing stages with SBX optimized algorithms, and enhancing users’ analysis workflows.”
Latest Parabricks benchmarks
In addition to new features and upgrades for each release, NVIDIA continuously works to improve benchmark performance across instruments, tools, and GPUs. Table 1 outlines the latest benchmarks on the most popular NVIDIA GPUs, including the newest Blackwell support in NVIDIA RTX PRO 6000.
Please note these benchmarks are from NVIDIA. Speeds can vary depending on the data set, GPU instance, and memory availability. Runtimes are for individual algorithms. When multiple algorithms are run together, pipelines can partially overlap and overall runtimes may be reduced.
| NVIDIA RTX PRO 6000 Newest Support | NVIDIA H100 SXM GPU | NVIDIA L4 GPU Lowest cost per sample | ||||
| 2 GPUs | 4 GPUs | 2 GPUs | 4 GPUs | 2 GPUs | 4 GPUs | |
| Giraffe(Paired-End) | 59.37 | 31.05 | 60.10 | 37.68 | 81.43 | 44.57 |
| DeepSomatic | 43.28 | 23.93 | 54.32 | 35.13 | 215.33 | 110.05 |
| Fq2bam(BWA-MEM) | 9.02 | 6.52 | 10.10 | 7.28 | 35.10 | 19.28 |
| rna_fq2bam(STAR) | 7.55 | 7.03 | 7.78 | 10.23 | 22.28 | 12.73 |
| fq2bam_meth(BWA-Meth) | 32.27 | 22.13 | 34.6 | 20.35 | 132.43 | 67.03 |
| DeepVariant | 4.67 | 2.75 | 7.60 | 5.90 | 13.83 | 8.03 |
| HaplotypeCaller | 8.33 | 3.83 | 8.63 | 4.17 | 14.2 | 7.03 |
| Mutect2 | 29.50 | 13.52 | 27.67 | 13.67 | 50.91 | 26.63 |
30x whole genome sequenced for Giraffe, fq2bam (BWA-MEM), DeepVariant, and Haplotype Caller with Illumina data.
50x tumor-normal whole genome sequenced for DeepSomatic and Mutect2 with Illumina data.
To benchmark fq2bam_meth (BWA-Meth), we downloaded whole genome bisulfite sequencing data from the ENCODE portal (Sloan et al. 2016) (https://www.encodeproject.org/) with the following identifiers: ENCSR890UQO (library ENCLB898WPW).
For rna_fq2bam (STAR) we used a FASTQ with 381,871,632 reads and a read length 98nt (SRA: SRR19653800).
Get started
NVIDIA Parabricks v4.5 provides improved functionality and reduced analysis time across industry-leading tools and offers support for the latest NVIDIA Blackwell architecture. Parabricks v4.5 is also accompanied by AI blueprints for genomics and single-cell analysis that lets users easily deploy and test NVIDIA Parabricks and NVIDIA RAPIDS.
Download NVIDIA Parabricks to get started with GPU-accelerated genomics analysis and join the conversation on the NVIDIA Parabricks Developer Forum.
Acknowledgments
Contributions and thanks to the Parabricks and RAPIDS teams. Particularly for this release, we want to acknowledge and thank a cross-functional team to ensure these new launches:
For NVIDIA RTX PRO 6000 Blackwell Server Edition benchmarks and Parabricks: Tong Zhu, Alejandro Chacon, Fabian Berressem, Daniel Puleri, and Yang Wang.
For AI blueprints with Brev.dev: Severin Dicks, Taurean Dyer, Gary Burnett, Alice Hsiung, and Andrew Walters.