Generative chemistry with AI has the potential to revolutionize how scientists approach drug discovery and development, health, and materials science and engineering. Instead of manually designing molecules with ��chemical intuition�� or screening millions of existing chemicals, researchers can train neural networks to propose novel molecular structures tailored to the desired properties.
]]>
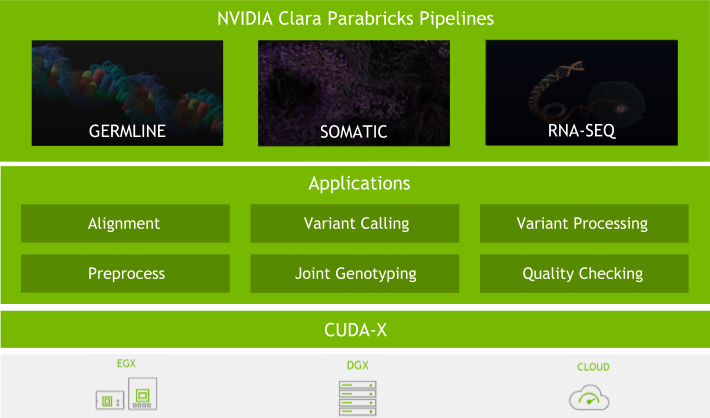
NVIDIA Parabricks is a scalable genomics analysis software suite that solves omics challenges with accelerated computing and deep learning to unlock new scientific breakthroughs. Released at NVIDIA GTC 2025, NVIDIA Parabricks v4.5 supports the growing quantity of data by including support for the latest NVIDIA GPU architectures, and improved alignment and variant calling with the��
]]>
Rare diseases are difficult to diagnose due to limitations in traditional genomic sequencing. Wolfgang Pernice, assistant professor at Columbia University, is using AI-powered cellular profiling to bridge these gaps and advance personalized medicine. At NVIDIA GTC 2024, Pernice shared insights from his lab��s work with diseases like Charcot-Marie-Tooth (CMT) and mitochondrial disorders.
]]>
NVIDIA Parabricks is a scalable genomics analysis software suite that solves omics challenges with accelerated computing and deep learning to unlock new scientific breakthroughs. NVIDIA Parabricks v4.4 introduces new features and functionality including accelerated pangenome graph alignment, as announced at the American Society of Human Genetics (ASHG) national meeting. The core new feature��
]]>
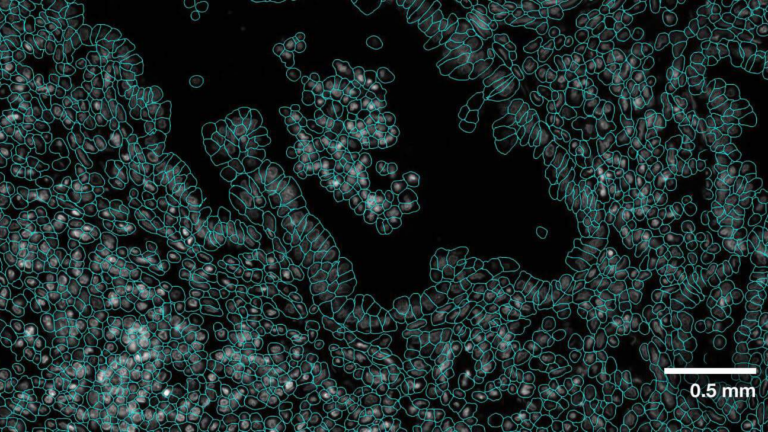
VISTA-2D is a new foundational model from NVIDIA that can quickly and accurately perform cell segmentation, a fundamental task in cell imaging and spatial omics workflows that is critical to the accuracy of all downstream tasks. The VISTA-2D model uses an image encoder to create image embeddings, which it can then turn into segmentation masks (Figure 1). The embeddings must contain��
]]>
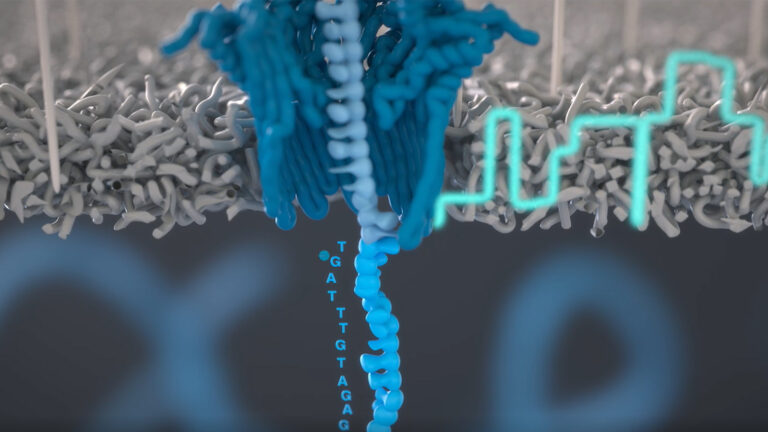
Geneformer is a recently introduced and powerful AI model that learns gene network dynamics and interactions using transfer learning from vast single-cell transcriptome data. This tool enables researchers to make accurate predictions about gene behavior and disease mechanisms even with limited data, accelerating drug target discovery and advancing understanding of complex genetic networks in��
]]>
NVIDIA Parabricks expands the NVIDIA emphasis on solving omics challenges with deep learning and continues accelerating genomics instruments. NVIDIA Parabricks v4.3.1, released at The European Society of Human Genetics (ESHG), introduces new functionality for variant calling in somatic data and upgrades to the latest versions of industry-leading tools. This release follows the recent Parabricks v4.
]]>
Missed GTC or want to replay your favorite training labs? Find it on demand with the NVIDIA GTC Training Labs playlist.
]]>
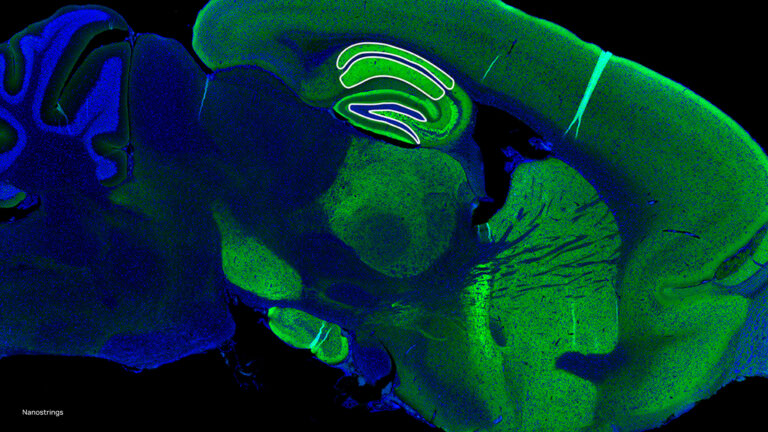
Genomics researchers use different sequencing techniques to better understand biological systems, including single-cell and spatial omics. Unlike single-cell, which looks at data at the cellular level, spatial omics considers where that data is located and takes into account the spatial context for analysis. As genomics researchers look to model biological systems across multiple omics at��
]]>
NVIDIA Parabricks v4.3 was released at NVIDIA GTC 2024, introducing new tooling and workflows that bring acceleration and the latest AI techniques to multiple omics data types. In addition to analyzing DNA and RNA, you can now also analyze methylation, single-cell, and spatial omics workloads at high speed and high accuracy with the power of GPUs and generative AI. Parabricks v4.3��
]]>
Learn how the Francis Crick Institute is using NVIDIA Clara Parabricks to enable key parts of TRACERx EVO, a new program that builds on the discoveries made in the world��s largest long-term lung study.
]]>
Parabricks version 4.2 has been released, furthering its mission to deliver unprecedented speed, cost-effectiveness, and accuracy in genomics sequencing analysis. The latest version delivers a newly accelerated workflow for Oxford Nanopore sequencing (in the featured image), enables Parabricks to be run on the latest NVIDIA GPUs, and furthers Parabricks�� accelerated deep learning variant calling��
]]>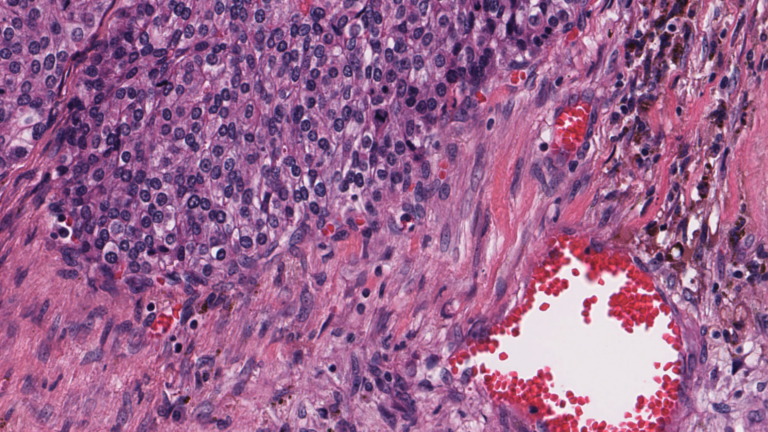
Digital pathology slide scanners generate massive images. Glass slides are routinely scanned at 40x magnification, resulting in gigapixel images. Compression can reduce the file size to 1 or 2 GB per slide, but this volume of data is still challenging to move around, save, load, and view. To view a typical whole slide image at full resolution would require a monitor about the size of a tennis��
]]>
Single-cell sequencing has become one of the most prominent technologies used in biomedical research. Its ability to decipher changes in the transcriptome and epigenome on a cell level has enabled researchers to gain valuable new insights. As a result, single-cell experiments have grown in size and complexity by a factor of over 100, with experiments involving more than 1 million cells becoming��
]]>
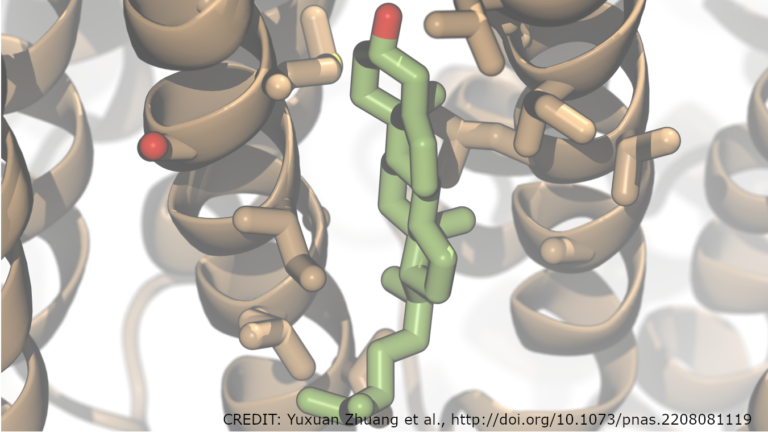
The human exome is key to understanding and treating genetic disorders and disease. Although the exome consists of just over 1% of the human genome, it also contains approximately 85% of known variants with significant disease-associated mutations. This is why whole-exome sequencing, involving the extraction and sequencing of these regions, is popular in clinical research and practice��
]]>
GPUs continue to get faster with each new generation, and it is often the case that each activity on the GPU (such as a kernel or memory copy) completes very quickly. In the past, each activity had to be separately scheduled (launched) by the CPU, and associated overheads could accumulate to become a performance bottleneck. The CUDA Graphs facility addresses this problem by enabling multiple GPU��
]]>
The upcoming 4.1 release of NVIDIA Parabricks, a suite of accelerated genomic analysis applications, goes further than ever before in accelerating sequencing alignment and increasing the accuracy of deep learning variant calling. The release includes a new workflow for PacBio long-read data, featuring an accelerated Minimap2 tool and Google��s DeepVariant for full GPU-enabled, end-to-end analysis��
]]>
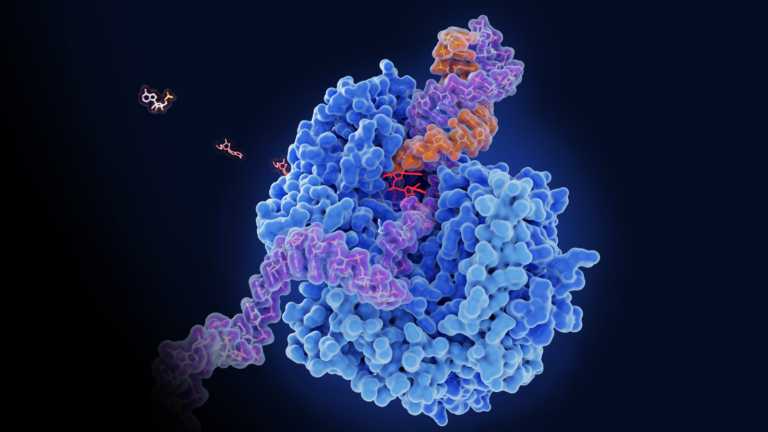
A collaboration between InstaDeep, the Technical University of Munich (TUM), and NVIDIA has led to the development of multiple super-computing scale foundation models for genomics. These models demonstrate state-of-the-art performance across many prediction tasks, such as promoter and enhancer site predictions. The joint team of researchers showed that large language models (LLMs) trained on��
]]>
Many organizations are using NVIDIA Clara Parabricks for fast human genome and exome analysis for large population projects, critically ill patients, clinical workflows, and cancer genomics projects. Their work aims to accurately and quickly identify disease-causing variants, keeping pace with accelerated next-generation sequencing as well as accelerated genomic analyses. Most recently��
]]>
Single-cell measurement technologies have advanced rapidly, revolutionizing the life sciences. We have scaled from measuring dozens to millions of cells and from one modality to multiple high dimensional modalities. The vast amounts of information at the level of individual cells present a great opportunity to train machine learning models to help us better understand the intrinsic link of cell��
]]>
The field of computational biology relies on bioinformatics tools that are fast, accurate, and easy to use. As next-generation sequencing (NGS) is becoming faster and less costly, a data deluge is emerging, and there is an ever-growing need for accessible, high-throughput, industry-standard analysis. At GTC 2022, we announced the release of NVIDIA Clara Parabricks v4.0��
]]>
Bioinformaticians are constantly looking for new tools that simplify and enhance genomic analysis pipelines. With over 60 tools, NVIDIA Clara Parabricks powers accurate and accelerated genomic analysis for germline and somatic workflows in research and clinical settings. Today, we announce the release of NVIDIA Clara Parabricks 3.8, featuring: The rapid increase in sequencing data��
]]>
Fast and cost-effective whole genome sequencing and analysis can bring answers quickly to critically ill patients suffering from rare or undiagnosed diseases. Recent advances in accelerated clinical sequencing, such as the world-record-setting DNA sequencing technique for rapid diagnosis, are bringing us one step closer to same-day, whole-genome genetic diagnosis in a clinical setting.
]]>
The following post provides a deep dive into some of the accomplishments and current focus of drug discovery and genomics work by NVIDIA. A leader in innovations within healthcare and life sciences, NVIDIA is looking to add AI, deep learning, simulation, and drug discovery researchers and engineers to the team. If what you read aligns with your career goals please review the current job postings.
]]>
The release of NVIDIA Clara Parabricks v3.6 last summer added multiple accelerated somatic variant callers and novel tools for annotation and quality control of VCF files to the comprehensive toolkit for whole genome and whole exome sequencing analysis. In the January 2022 release of Clara Parabricks v3.7, NVIDIA expanded the scope of the toolkit to new data types while continuing to improve��
]]>
The newest release of GPU-powered NVIDIA Clara Parabricks v3.7 includes updates to germline variant callers, enhanced support for RNA-Seq pipelines, and optimized workflows for gene panels. With now over 50 tools, Clara Parabricks powers accurate and accelerated genomic analysis for gene panels, exomes, and genomes for clinical and research workflows. To date��
]]>
Single-cell genomics research continues to advance drug discovery for disease prevention. For example, it has been pivotal in developing treatments for the current COVID-19 pandemic, identifying cells susceptible to infection, and revealing changes in the immune systems of infected patients. However, with the growing availability of large-scale single-cell datasets, it��s clear that computing��
]]>
The release of NVIDIA Clara Parabricks v3.6 brings new applications for variant calling, annotation, filtering, and quality control to its suite of powerful genomic analysis tools. Now featuring over 33 accelerated tools for every stage of genomic analysis, NVIDIA Clara Parabricks provides GPU-accelerated bioinformatic pipelines that can scale for any workload. As genomes and exomes are��
]]>
Here are the latest resources and news for healthcare developers from GTC 21, including demos and specialized sessions for building AI in drug discovery, medical imaging, genomics, and smart hospitals. Learn about new features now available in NVIDIA Clara Train 4.0, an application framework for medical imaging that includes pre-trained models, AI-assisted annotation, AutoML��
]]>
The Human Genome Center (HGC) at University of Tokyo announced a new genomics platform to accelerate genomic analysis by 40X compared to a CPU-based environment, utilizing NVIDIA Clara Parabricks Pipelines genomics software. The genomic analysis will run on SHIROKANE, HGC��s fastest supercomputer for life sciences in Japan, and powered by NVIDIA DGX A100. The platform will be available to users on��
]]>
Using the highest-resolution images of a single DNA molecule captured to date, researchers in the U.K. discovered that coiled strands of genetic material twist and writhe while crammed in a cell. This previously unobserved movement was simulated on GPU-based systems including JADE, a University of Oxford supercomputer made up of NVIDIA DGX systems. Published in Nature Communications��
]]>
NVIDIA recently released NVIDIA Clara Parabricks Pipelines version 3.5, adding a set of new features to the software suite that accelerates end-to-end genome sequencing analysis. With the release of v3.5, Clara Parabricks Pipelines now provides acceleration to Google��s DeepVariant 1.0, in addition to a suite of existing DNA and RNA tools. The addition of DeepVariant to Parabricks Pipelines��
]]>
This week at GTC 2020, synthetic biology startup Synvivia showcased protein switches being developed to control engineered organisms and aid in drug discovery for COVID-19. The full session is available in the GTC catalog to view on-demand. Synvivia uses GPU-accelerated molecular dynamics simulations to design protein molecule interactions and was able to observe potential additive effects of��
]]>
Whole genome sequencing has become an important and foundational part of genomic research, enabling researchers to identify genetic signatures associated with diseases, differentiate sequencing errors from biological signals, and better characterize the genomes of various organisms. With the ongoing COVID-19 pandemic threatening the globe, characterizing, and understanding genomes is now more��
]]>
The human body is made up of nearly 40 trillion cells, of many different types. Recent advances in experimental biology have made it possible to explore the genetic material of single cells. With the birth of this new field of single-cell genomics, scientists can now probe the DNA and RNA of individual cells in the human body. Single-cell genomic analysis has identified new types of cells in��
]]>
To help accelerate natural language processing in biomedicine, Microsoft Research developed a BERT-based AI model that outperforms previous biomedicine natural languag processing (NLP) methods. The work promises to help researchers rapidly advance research in this field. The model, built on top of Google��s BERT, can classify documents, extract medical information��
]]>
The world of healthcare is under a giant spotlight these days. In these unprecedented times, everyone is focusing on keeping loved ones safe and contributing to the community as much as possible. It is amazing to see how the community is uniting in the past two months, which has led to a global, rapid emergence of SARS-CoV-2 projects across the life sciences. At NVIDIA��
]]>
Genomics analysis is playing a key role in COVID-19 studies, as the data from sequencing projects is helping researchers better understand and characterize the coronavirus. With NVIDIA Parabricks, researchers can integrate GPU power into existing genomics workflows and rapidly sequence fragments to enhance research. In the COMPUTE4COVID webinar series, we discuss in detail how sequencing��
]]>
Starting now, NVIDIA will provide a free 90-day license to Parabricks to any researcher in the worldwide effort to fight the novel coronavirus. Based on the well-known Genome Analysis Toolkit, Parabricks uses GPUs to accelerate by as much as 50x the analysis of sequence data. If you have access to NVIDIA GPUs, fill out this form to request a Parabricks license. For researchers working with��
]]>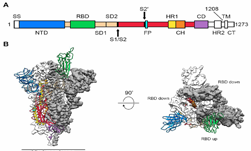
To help in the development of a treatment or vaccine for COVID-19, University of Texas at Austin and National Institute of Health (NIH) researchers recently achieved a critical breakthrough��creating the first 3D, atomic-scale map of the virus. ��2019-nCoV makes use of a densely glycosylated spike (S) protein to gain entry into host cells,�� the researchers stated in a newly published Science��
]]>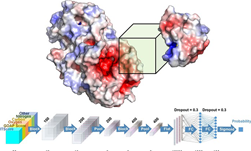
To help better understand how proteins interact in the body, Purdue University researchers developed a deep learning-based approach for modeling protein interactions, a first for AI the researchers say. The work has the potential to help fight various diseases and to design better drugs that directly target protein interactions. ��Our work represents a major advancement in the field of��
]]>
The Clara Genomics SDK has been upgraded with high performance analysis algorithms for long read sequencing and early access to deep learning-based processing of short read ATAC sequencing. Features: GPU Accelerated Racon The Racon consensus module for genome assembly enabled GPU- accelerated polishing of long reads in v1.4 with the integration of the cudaPOA module in��
]]>
Today, NVIDIA is releasing the Clara Genomics Analysis SDK �C an open source toolkit for biological sequence analysis, that is part of Clara Genomics. The last few years have seen a revolution in genome sequencing. New high-throughput sequencing techniques such as those developed by Oxford Nanopore, PacBio and Illumina have led to increases in throughput and quality as well as large reductions��
]]>
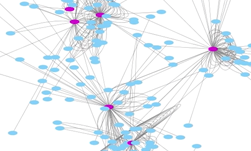
A new research paper published in the journal Nature Genomics shows how a Princeton University-led team developed a deep learning-based method for searching the human genome for mutations, including their associations with diseases such as autism. ��Many mutations in DNA that contribute to disease are not in actual genes but instead lie in the 99% of the genome once considered ��junk.
]]>
The opioid crisis has become one of the worst epidemics in U.S. history, claiming thousands of lives every year. In 2016, over 50,000 people died from opioid overdoses, and in 2017 it is predicted that over 72,000 people died from drug overdoses. To help get a better understanding of how opioids interact with human genes, researchers from the Oak Ridge National Laboratory have enlisted the help��
]]>
At GTC Europe in Munich, Germany, U.K. startup Oxford Nanopore Technologies just introduced the MiniIT hand-held AI supercomputer, powered by NVIDIA AGX, which delivers a low-cost, real-time DNA and RNA sequencer. ��MinIT is the perfect, powerful companion to MinION, the only portable real-time DNA sequencer,�� said Gordon Sanghera, CEO of Oxford Nanopore. ��As data streams faster from our��
]]>
The Biowulf supercomputer at the National Institutes of Health (NIH) received an upgrade with the addition of 144 NVIDIA GPUs to help researchers discover new cures and save lives. According to CSRA, the system integrator and service company responsible for the installation: ��The second stage of computing power announced today will enable NIH researchers to make important advances in biomedical��
]]>
With our planet getting warmer and warmer, and carbon dioxide levels steadily creeping up, companies are using deep learning to help cope with the effects that climate change is having on their crops. An article on MIT Technology Review highlights PEAT, a German company using CUDA, TITAN X GPUs and the cuDNN-accelerated Caffe deep learning framework to provide farmers with a plant disease and��
]]>
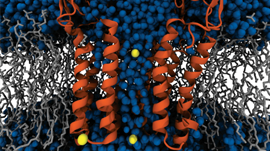
Understanding the critical genetic traits of agricultural crops will help develop crops faster in order to handle the world��s population growth and climate change. In a collaboration between Biochemist Alex Feltus and Computer Engineering PhD student Karan Sapra, the Clemson University researchers have created an interactive visualization tool using NVIDIA GPUs that allows them to look at complex��
]]>
Erik Lindahl, Professor of Biophysics at Stockholm University, talks about using the GPU-accelerated GROMACS application to simulate protein dynamics. This approach helps researchers learn how to design better drugs, combat alcoholism and understand how certain diseases occur. Lindahl mentions they started using CUDA in their work nearly five years ago and now 90% of their computational resources��
]]>
Cancer kills almost 600,000 people each year in the U.S. alone. Researchers from the University of Toronto are advancing computational cancer research by developing a ��genetic interpretation engine�� �C a GPU-powered, deep learning method for identifying cancer-causing mutations. Under its Compute the Cure initiative, the NVIDIA Foundation awarded the team a $200,000 research grant to further��
]]>
The list of the world��s TOP500 supercomputers was just published and shows the extent to which accelerated systems are shaping the future of the industry. For the first time, more than 100 accelerated systems are on the list of the world��s 500 most powerful supercomputers, and 70 of these are Tesla-based supercomputers �C including 23 of the 24 new systems on the list which is nearly 50 percent��
]]>
NVBIO is an open-source C++ template library of high performance parallel algorithms and containers designed by NVIDIA to accelerate sequence analysis and bioinformatics applications. NVBIO has a threefold focus: We built NVBIO because we believe only the exponentially increasing parallelism of many-core GPU architectures can provide the immense computational capability required by the��
]]>