Pharmaceutical research demands fast, efficient simulations to predict how molecules interact, speeding up drug discovery. Jiqun Tu, a senior developer technology engineer at NVIDIA, and Ellery Russell, tech lead for the Desmond engine at Schr?dinger, explore advanced GPU optimization techniques designed to accelerate molecular dynamics simulations. In this NVIDIA GTC 2024 session��
]]>
NVIDIA Nsight Systems is a comprehensive tool for tracking application performance across CPU and GPU resources. It helps ensure that hardware is being efficiently used, traces API calls, and gives insight into inter-node network communication by describing how low-level metrics sum to application performance and finding where it can be improved. Nsight Systems can scale to cluster-size��
]]>
GPUs continue to get faster with each new generation, and it is often the case that each activity on the GPU (such as a kernel or memory copy) completes very quickly. In the past, each activity had to be separately scheduled (launched) by the CPU, and associated overheads could accumulate to become a performance bottleneck. The CUDA Graphs facility addresses this problem by enabling multiple GPU��
]]>
GROMACS, a scientific software package widely used for simulating biomolecular systems, plays a crucial role in comprehending important biological processes important for disease prevention and treatment. GROMACS can use multiple GPUs in parallel to run each simulation as quickly as possible. Over the past several years, NVIDIA and the core GROMACS developers have collaborated on a series of��
]]> ]]>
]]>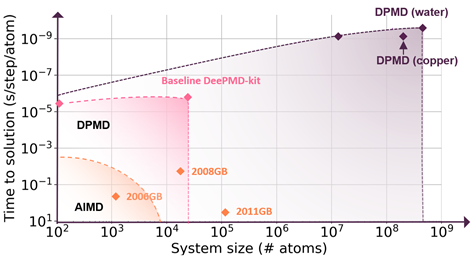
Molecular simulation communities have faced the accuracy-versus-efficiency dilemma in modeling the potential energy surface and interatomic forces for decades. Deep Potential, the artificial neural network force field, solves this problem by combining the speed of classical molecular dynamics (MD) simulation with the accuracy of density functional theory (DFT) calculation.1 This is achieved by��
]]>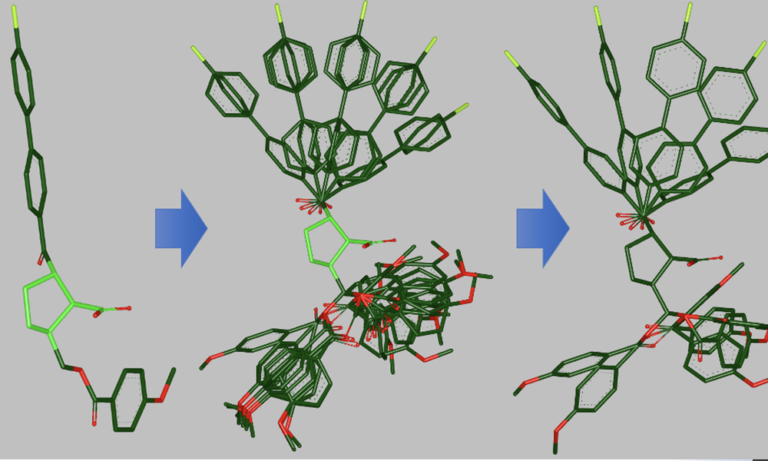
Computational molecular design involves compute-intense calculations that require exceptional processing power. Whether working in pharmaceuticals, biotechnology, agrochemicals, or the fragrance industry, researchers are oftentimes dealing with datasets that encompass millions to billions of compounds. Until recently, this required that companies invest in expensive��
]]>
The latest breakthroughs in graphics technologies are elevating workflows across industries��and you can experience it all at the NVIDIA GTC, begins November 8. There are several GTC sessions for professional content creators, engineers, and developers looking to explore new tools and techniques accelerated by NVIDIA. We will showcase how NVIDIA is powering real-time ray tracing��
]]>
GROMACS, a simulation package for biomolecular systems, is one of the most highly used scientific software applications worldwide, and a key tool in understanding important biological processes including those underlying the current COVID-19 pandemic. In a previous post, we showcased recent optimizations, performed in collaboration with the core development team, that enable GROMACS to��
]]>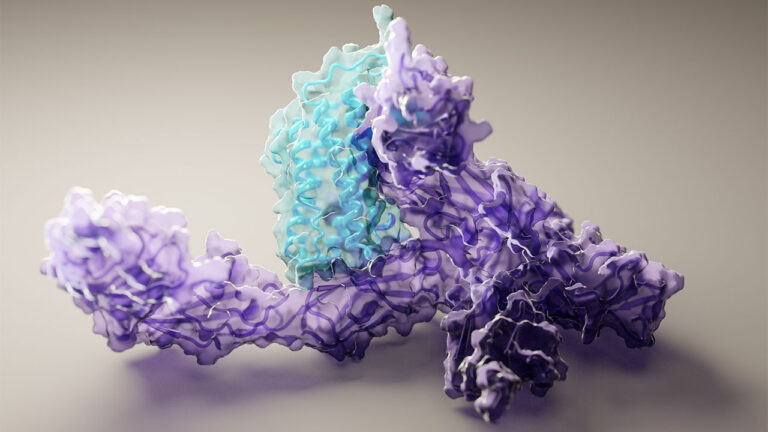
Solving a mystery that stumped scientists for decades, last November a group of computational biologists from Alphabet��s DeepMind used AI to predict a protein��s structure from its amino acid sequence. Not even a year later, a new study offers a more powerful model, capable of computing protein structures in as little as 10 minutes, on one gaming computer. The research��
]]>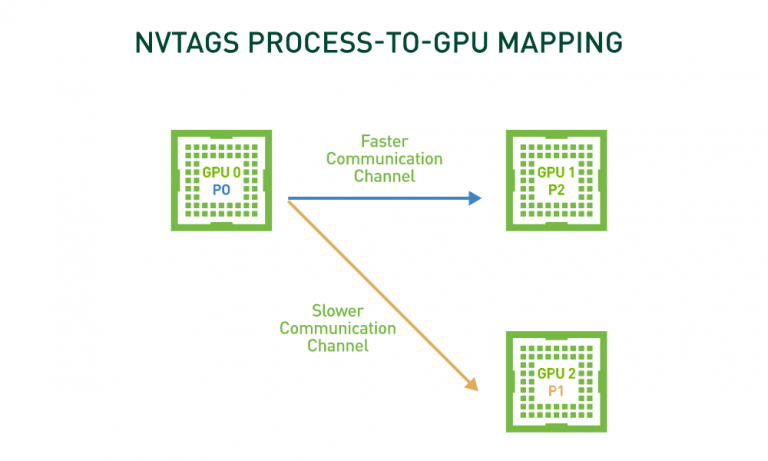
Many GPU-accelerated HPC applications spend a substantial portion of their time in non-uniform, GPU-to-GPU communications. Additionally, in many HPC systems, different GPU pairs share communication links with varying bandwidth and latency. As a result, GPU assignment can substantially impact time to solution. Furthermore, on multi-node / multi-socket systems, communication performance can degrade��
]]>
A container is a portable unit of software that combines the application and all its dependencies into a single package that is agnostic to the underlying host OS. In a high-performance computing (HPC) environment, containers remove the need for building complex environments or maintaining environment modules, making it easy for researchers and systems administrators to deploy their HPC��
]]>
From weather forecasting and energy exploration, to computational chemistry and molecular dynamics, NVIDIA compute and networking technologies are optimizing nearly 2,000 applications across a broad-range of scientific domains and industries. By leveraging GPU-powered parallel processing, users can accelerate advanced, large-scale applications efficiently and reliably, paving the way to scientific��
]]>
Simulations by Lawrence Livermore National Laboratory researchers have uncovered a new mechanism for freezing in metals, advancing scientists�� understanding of nucleation, the process of gases or liquids cooling into crystalline solids. Run on 256 NVIDIA Tensor Core GPUs on the Lassen supercomputer, the simulations modeled how heated copper solidifies, providing atomic-scale insights into the��
]]>
Wouldn��t it be amazing if you could create beautiful and immersive scientific visualizations of large and dynamic simulations like Folding@Home��s simulation of COVID-19 spikes? In this post, we share our recipe to show that you can use NVIDIA Omniverse to create powerful cinematic visualizations from scientific data. The beginning of any such project starts with trajectory data.
]]>
��Meet the Researcher�� is a monthly series in which we spotlight different researchers in academia who are using NVIDIA technology to accelerate their work. This month, we spotlight Gregory A. Voth, Distinguished Professor at the Department of Chemistry, The University of Chicago. Voth received a Ph.D. in Theoretical Chemistry from the California Institute of Technology in 1987 and was an IBM��
]]>
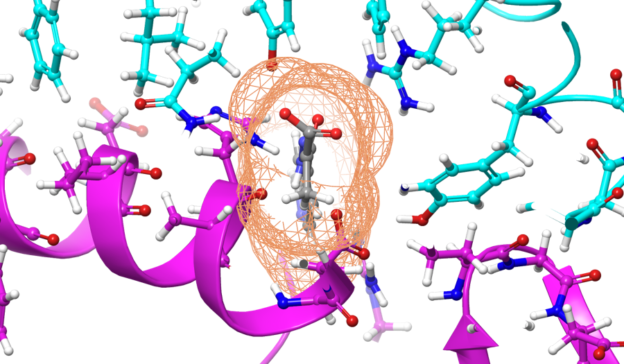
This week at GTC 2020, synthetic biology startup Synvivia showcased protein switches being developed to control engineered organisms and aid in drug discovery for COVID-19. The full session is available in the GTC catalog to view on-demand. Synvivia uses GPU-accelerated molecular dynamics simulations to design protein molecule interactions and was able to observe potential additive effects of��
]]>
As the world battles to reach a scientific breakthrough in the fight against COVID-19, scientists are turning to computing resources to accelerate their research. To help make the process for scientists more accessible, we��re spotlighting a few of the GPU-accelerated applications that developers can use right now in the fight against this virus. Applications like AMBER, GROMACS, NAMD��
]]>
To help tackle COVID-19, the long-running Folding@Home program, a distributed computing project for simulating protein dynamics, hit a breakthrough by achieving more than an exaflop of processing power. That��s more than 1,000,000,000,000,000,000 operations per second, all through crowdsourcing efforts. For comparison, Summit, the world��s fastest supercomputer, which is powered by more than 27��
]]>
To help respond to the COVID-19 coronavirus outbreak, researchers at the Oak Ridge National Laboratory (ORNL) are using the world��s fastest supercomputer to identify compounds that may effectively combat the virus. Using Summit, which is powered by 9,216 IBM Power9 CPUs and over 27,000 NVIDIA V100 Tensor Core GPUs, the researchers identified 77 small-molecule drug compounds that are likely��
]]>
GROMACS��one of the most widely used HPC applications�� has received a major upgrade with the release of GROMACS 2020. The new version includes exciting new performance improvements resulting from a long-term collaboration between NVIDIA and the core GROMACS developers. As a simulation package for biomolecular systems, GROMACS evolves particles using the Newtonian equations of motion.
]]>
Whether you are a HPC research scientist, application developer, or IT staff, NVIDIA has solutions to help you use containers to be more productive. NVIDIA is enabling easy access and deployment of HPC applications by providing tuned and tested HPC containers on the NGC registry. Many commonly used HPC applications such as NAMD, GROMACS, and MILC are available and ready to run just by downloading��
]]>
NVIDIA GPUs power the world��s fastest supercomputer, and 20 of the 100 most powerful supercomputing clusters in the world are also powered by them too. If you follow NVIDIA closely, you are probably not surprised by this, but in a new article published in Nature this week, the leading scientific publication explains why so many researchers and developers are using NVIDIA GPUs to accelerate their��
]]>
Researchers from Google, along with collaborators from academia, announced today they developed a deep learning-based system for identifying protein crystallization, achieving a 94 percent accuracy rate. Protein crystallization plays a vital role in the drug discovery process, helping determine the shape of cells. The work has the potential to further the drug discovery process by making it��
]]>
Physicists from more than a dozen institutions used the power of the GPU-accelerated Titan Supercomputer at Oak Ridge National Laboratory to calculate a subatomic-scale physics problem of measuring the lifetime of neutrons. The study, published in Nature this week, achieves groundbreaking precision and provides the research community with new data that could aid in the search for dark matter and��
]]>
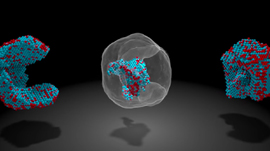
For the first time ever, chemists from the University of California, San Diego designed a two-dimensional protein crystal simulation that toggles between states of varying porosity and density. The research could help scientists create new materials for water purification, renewable energy, breakthroughs in medicine, drug development, and many more possible applications. The work of combining��
]]>
Scientists used an extremely high-resolution transmission electron microscope to capture 2D projections of the nanoparticle��s structure, and used an algorithm to stitch those together into a 3D reconstruction. The unprecedented detail sheds light on the material��s properties at the single-atom level and the insights gained from the particle��s structure could lead to new ways to improve its��
]]>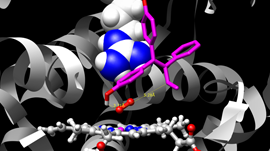
Thomas Cheatham, professor of Medicinal Chemistry and the director of research computing at University of Utah shares how they��re using the GPU-accelerated Blue Waters supercomputer and NVLink to compute the interactions of atoms that can lead to drug design and materials design. ��The GPUs have been really helpful, because we��ve optimized our codes (AMBER, a package of programs for molecular��
]]>
Gil Speyer, Senior Postdoctoral Fellow at the Translational Genomics Research Institute (TGen) shares how NVIDIA technology is accelerating the computer processing of transcriptomes from thousands of cells gleaned from patient tumor samples. Using NVIDIA Tesla K40 GPUs and CUDA, the scientists developed a statistical analysis tool called EDDY (evaluation of differential dependency) that examines��
]]>
Accelerated by 200 Tesla K80 GPUs, the Laconia Supercomputer was recently unveiled at the Institute for Cyber-Enabled Research at Michigan State University. Named after a region in Greece that was home to the original Spartans, the mascot of Michigan State University, the supecomputer ranks in the TOP500 fastest computers in the world and is projected to be in the top 6%
]]>
A team lead by Cornell University researchers are using the Titan Supercomputer at Oak Ridge National Laboratory to study mechanisms of sodium-powered transporters in cell-to-cell communications. Harel Weinstein��s lab at the Weill Cornell Medical College of Cornell University have constructed complex 3D molecular models of a specific family of neurotransmitter transporters called neurotransmitter��
]]>
The Facebook Artificial Intelligence Research (FAIR) lab announced a new Research Partnership Program to spur advances in Artificial Intelligence and machine learning �� Facebook will be giving out 25 servers powered with GPUs, free of charge. The first recipient to receive 32 GPUs in four GPU servers is Klaus-Robert M��ller of TU Berlin. ��Dr. M��ller will receive four GPU servers that will enable��
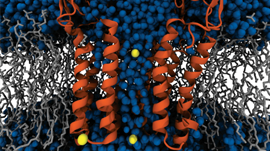
]]>
Erik Lindahl, Professor of Biophysics at Stockholm University, talks about using the GPU-accelerated GROMACS application to simulate protein dynamics. This approach helps researchers learn how to design better drugs, combat alcoholism and understand how certain diseases occur. Lindahl mentions they started using CUDA in their work nearly five years ago and now 90% of their computational resources��
]]>
Research Area Specialist Dr. Joshua A. Anderson at University of Michigan was an early user of GPU computing technology. He began his career developing software on the first CUDA capable GPU, and now runs simulations on one of the world��s most powerful supercomputers. His ��contributions to the development and dissemination of the open source, GPU-enabled molecular simulation software, HOOMD-blue��
]]>
Dr. Joshua A. Anderson is a Research Area Specialist at the University of Michigan who was an early user of GPU computing technology. He began his career developing software on the first CUDA capable GPU and now runs simulations on one of the world��s most powerful supercomputers. Anderson��s ��contributions to the development and dissemination of the open source, GPU-enabled molecular simulation��
]]>
Dr. Diego Rossinelli, an ETH Zurich researcher and 2015 Gordon Bell Finalist, shares how they are relying on CUDA and Tesla GPUs to track down tumor cells that are markers of metastatic cancer. Watch Diego��s talk ��18,688 K20X��s Running after a Tumor Cell�� in the NVIDIA GPU Technology Theater at SC15: Watch Now Read more about his research at http://nvda.ly/UR7sQ Share your GPU��
]]>
Oliver Laslett, Post-Graduate Researcher at University of Southampton discusses how magnetic nanotechnology can be used to improve bio-medicine. Share your GPU-accelerated science with us: http://nvda.ly/Vpjxr Watch more scientists and researchers share how accelerated computing is #thepathforward: Watch Now ��
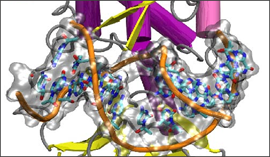
]]>
Ross Walker, AMBER developer at San Diego Computing Super Center and University of California, San Diego discusses how they are accelerating molecular dynamics to transform drug design. Read more about his research at http://bit.ly/1lx4A80 Share your GPU-accelerated science with us: http://nvda.ly/Vpjxr Watch more scientists and researchers share how accelerated computing is #
]]>
Jim Phillips, Senior Research Programmer at University of Illinois at Urbana-Champaign is using the Tesla-accelerated supercomputers, Titan and Blue Waters for his parallel molecular dynamics code, NAMD, designed for high-performance simulation of large biomolecular systems. Watch Jim��s talk ��Petascale Biomolecular Simulation with NAMD on Titan, Blue Waters, and Summit�� in the��
]]>
For the past 30 years, users of the San Diego Supercomputer Center (SDSC) systems have achieved major scientific breakthroughs spanning many domains, from earth sciences and biology to astrophysics, bioinformatics, and health IT. A few milestones include: 1987: Scientists take a major step in the new arena of rational drug design, determining the relative free energies of binding for different��
]]>
HPC looks very different today than it did when I was a graduate student in the mid-90s. Today��s supercomputers are many orders of magnitude faster than the machines of the 90s, and GPUs have helped push arithmetic performance on several leading systems to stratospheric levels. Unfortunately, the arithmetic performance wrought by two decades of supercomputer design has created tremendous I/
]]>
Our Spotlight is on Dr. Michela Taufer, Associate Professor at the University of Delaware. Michela heads the Global Computing Lab (GCLab), which focuses on high performance computing (HPC) and its application to the sciences. Her research interests include software applications and their advanced programmability in heterogeneous computing (i.e., multi-core platforms and GPUs)��
]]>